ABI 3500xl genetic analyzer information
The facility DNA sequencing system was upgraded in January 2021 from the ABI 3130xl to the ABI 3500xl. Following is some information on the system:
Protocols
- Sequencing reaction protocol for BigDye: BigDye Terminator v3.1 Cycle Sequencing Kit Protocol (Applied Biosystems protocol)
Recommended DNA purification kits
- Qiagen DNA purification kits
- Other purification kits may be suitable—please contact the facility to discuss
Notes on analysis of results with Sequence Analysis Software V7.0
- KB basecaller V1.4.1:
- A novel basecaller algorithm that performs basecalling for pure and mixed base calls.
- Generation of quality values (QV) to provide basecall accuracy information for pure and mixed base calls.
- QV = -10 log10 (Pe)Where Pe is the probability of error
- The KB basecaller generates QVs from 1 to 99, and the QVs are optionally displayed as bars above each base in the Sequences and Electropherograms views.
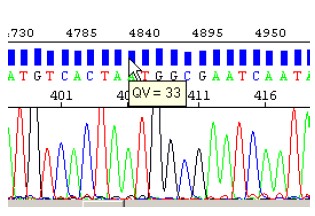
- Clear range is the region of a DNA sequence that remains after excluding the low quality or error-prone sequence at both 5’and 3’ ends.
- Multiple formats of output sequence files:
- Analyzed ABI files (.ab1)
- Text file in ABI or Fasta (.seq)
- Phred (.phd.1) files
- Standard chromatogram format (.scf) files
- Acrobat (.pdf)
Generally we provide ABI and Fasta files, but if clients require other type of the sequence files, we can re-analyze data, and then send phred or chromatogram files.
Software for sequence file analysis
- ThermoFisher Connect online Data Analysis Apps. Click here to connect.
- Finch TV to view, edit, print, and blast your sequence data.
- ApE-A Plasmid Editor, to view, edit, and analyze data, including alignment of multiple sequences.
- Sequence Analysis Sofware from ABI to edit and view both analyzed and raw sequence data, clear range at 5’ and 3’ ends, QVs and QV bars for the sequences, and export traces in multiple formats including pdf, phd.1, seq etc.
Feedback
If you have any suggestions for additions or edits to this page, please contact the facility.
Return to DNA Sequencing Core Facilities main page.